Research
Variation in pathogenicity and virulence among Group B Streptococcus (GBS)
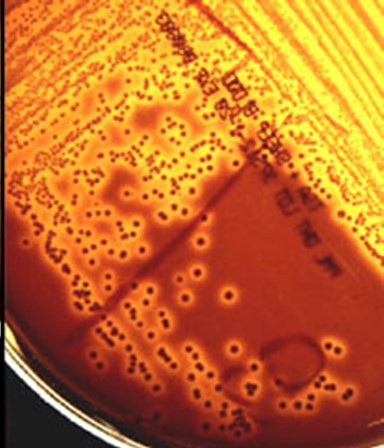
GBS hemolysis
Photo credit: Rebekah Sloup
GBS is an opportunistic pathogen that can be transmitted vertically to babies in utero or following childbirth, leading to preterm birth, stillbirth, sepsis or meningitis. Through our studies, we have demonstrated that most neonatal isolates belong to a specific genotype that differs from prenatal colonizing isolates and that strains of this genotype have unique traits that can impact pathogenesis, host-pathogen interactions, and disease outcomes.
Genomic epidemiology of foodborne pathogens
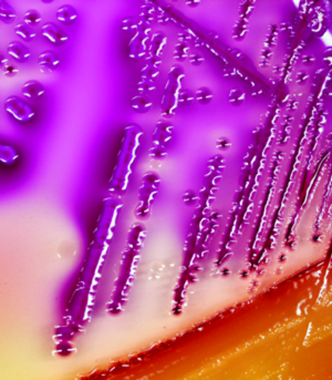
STEC on selective media
Photo credit: Rebekah Sloup
Shiga toxin-producing Escherichia coli (STEC), Campylobacter jejuni, and Salmonella spp. frequently cause foodborne outbreaks of diarrheal disease. Our studies have focused on using genomics to characterize these pathogens from humans with clinical infections for comparison to those from important reservoirs, like cattle. We have observed the emergence of antibiotic resistance, classified genetic mechanisms of resistance, and uncovered phenotypes and traits that are important for persistence in reservoir hosts and virulence in humans. We also have identified epidemiological risk factors for more severe infections as well as those caused by drug-resistant strains.
Impact of disease and antibiotic use on the gut microbiome
Another research focus has been on examining how various perturbations affect the gut microbiome using 16S rRNA sequencing, metagenomics, and metabolomics. Our analysis of microbiota and microbiome profiles in patients with diarrheal disease (cases) and their healthy family members (controls) detected differences in the abundance of certain microbes across groups and showed that increases in some species are linked to poor clinical outcomes. We further showed an increase in the abundance and diversity of antibiotic resistance genes in the patients and discovered that critically important resistance genes persisted even after they recovered from their infections. We have conducted similar studies in dairy cattle and have demonstrated that antibiotic use contributes to the persistence of specific antibiotic resistance genes even after treatment cessation.